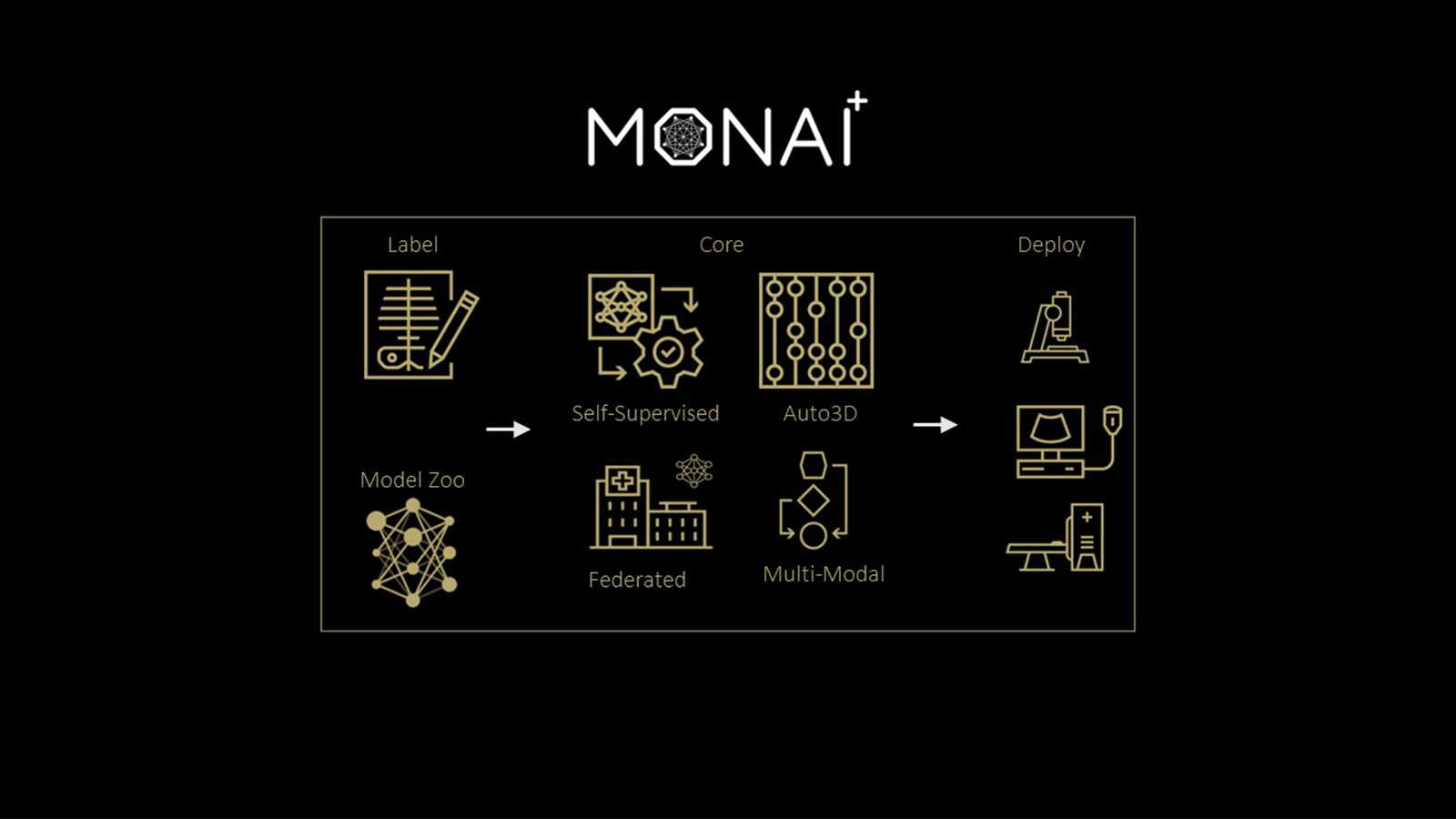
The Medical Open Network for AI (MONAI), is a freely available, community-supported, PyTorch-based framework for deep learning in healthcare imaging. It provides domain-optimized, foundational capabilities for developing a training workflow.
Building upon the GTC 2020 alpha release announcement back in April, MONAI has now released version 0.2 with new capabilities, examples, and research implementations for medical imaging researchers to accelerate the pace of innovation for AI development. For more information, see NVIDIA and King’s College London Announce MONAI Open Source AI Framework for Healthcare Research.
Why MONAI research?
MONAI research is a submodule in the MONAI codebase. The goal is to showcase the implementation of research prototypes and demonstrations from recent publications in medical imaging with deep learning. The research modules are regularly reviewed and maintained by the core developer team. Reusable components identified from the research submodule are integrated into the MONAI core module, following good software engineering practices.
Along with the flexibility and usability of MONAI, we envision MONAI research as a suitable venue to release the research code, increase the research impact, and promote open and reproducible research. Like all the other submodules in MONAI, we welcome contributions in the forms of comments, ideas, and code.
In this post, we discuss the research publications that now have been included with a MONAI-based implementation, addressing advanced research problems in medical image segmentation. MONAI is not intended for clinical use.
- COPLE-Net: COVID-19 Pneumonia Lesion Segmentation Network
- LAMP: Large Deep Nets with Automated Model Parallelism for Image Segmentation
COPLE-Net: COVID-19 Pneumonia Lesion Segmentation Network
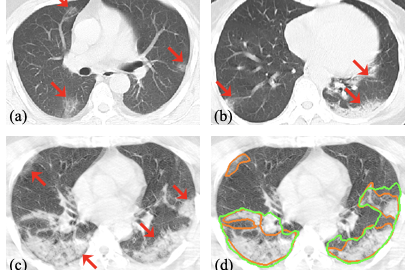
Segmentation of pneumonia lesions from CT scans of COVID-19 patients is important for accurate diagnosis and follow-up. In a recent paper, the leading author, Guotai Wang from University of Electronic Science and Technology of China, and the team propose to use deep learning to automate this task. For more information, see A Noise-robust Framework for Automatic Segmentation of COVID-19 Pneumonia Lesions from CT Images.
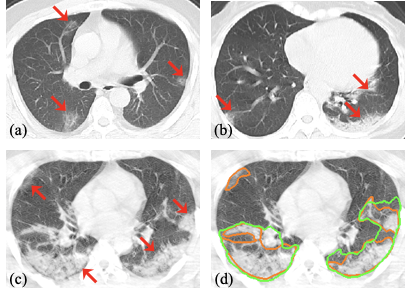
Acquiring a large set of accurate pixel-level annotations of the pneumonia lesions during the outbreak of COVID-19 is challenging. This research mainly deals with learning from noisy labels for the segmentation task.
One of the key innovations of the research is an enhanced deep convolutional neural network architecture. This architecture has the following features:
- It uses a combination of max-pooling and average pooling to reduce information loss during downsampling.
- It employs bridge layers to alleviate the semantic gap between features in the encoder and decoder.
- It employs an ASPP module at the bottleneck to better deal with lesions at multiple scales.
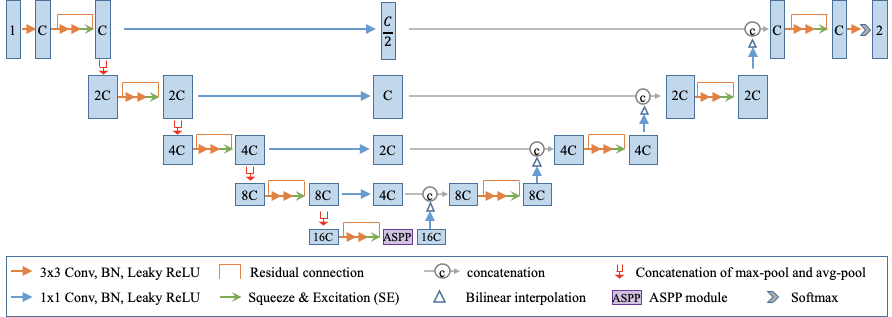
The novel architecture is made available in MONAI. The key network components, such as MaxAvgPool and SimpleASPP, could be conveniently integrated into other deep learning pipelines:
from monai.networks.blocks import MaxAvgPool, SimpleASPP max_avg_pool = MaxAvgPool(spatial_dims=spatial_dims, kernel_size=2) aspp = SimpleASPP(spatial_dims, ft_chns[4], int(ft_chns[4] / 4), kernel_sizes=[1, 3, 3, 3], dilations=[1, 2, 4, 6])
The image preprocessing pipeline and pretrained model loading could be done in a few Python commands with MONAI:
images = sorted(glob(os.path.join(IMAGE_FOLDER, "case*.nii.gz")))
val_files = [{"img": img} for img in images]
# define transforms for image and segmentation
infer_transforms = Compose(
[
LoadNiftid("img"),
AddChanneld("img"),
Orientationd("img", "SPL"),
ToTensord("img"),
]
)
test_ds = monai.data.Dataset(data=val_files, transform=infer_transforms)
# sliding window inference need to input one image in every iteration
data_loader = torch.utils.data.DataLoader(
test_ds, batch_size=1, num_workers=0, pin_memory=torch.cuda.is_available()
)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = CopleNet().to(device)
model.load_state_dict(torch.load(MODEL_FILE)["model_state_dict"])
The PyTorch users would benefit from the MONAI medical image preprocessors and domain specific network blocks. At the same time, the code shows the compatibility of MONAI modules and the PyTorch native objects such as torch.utils.data.DataLoader, thus facilitating the easy adoption of MONAI modules in general PyTorch workflows.
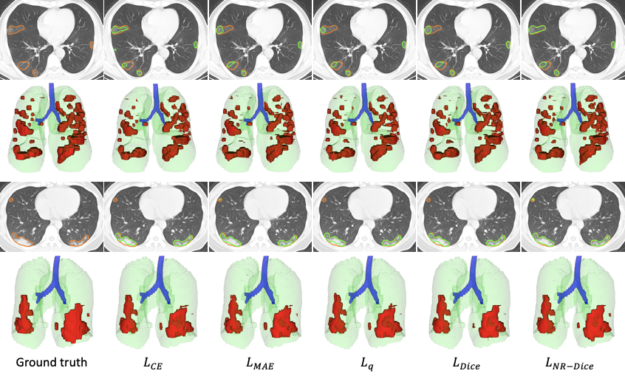
In the scenario of learning from noisy labels for COVID-19 pneumonia lesion segmentation, the experimental results of the COPLE-Net demonstrate that the new architecture can achieve higher performance than state-of-the-art CNNs.
LAMP: Large Deep Nets with Automated Model Parallelism for Image Segmentation
Deep learning models are becoming larger because increases in model size can offer significant accuracy gain. Through automated model parallelism, it is feasible to train large deep 3D ConvNets with a large input patch, even the whole image. For more information about the possibility of the automated model parallelism for 3D U-Net for medical image segmentation tasks, see LAMP: Large Deep Nets with Automated Model Parallelism for Image Segmentation.
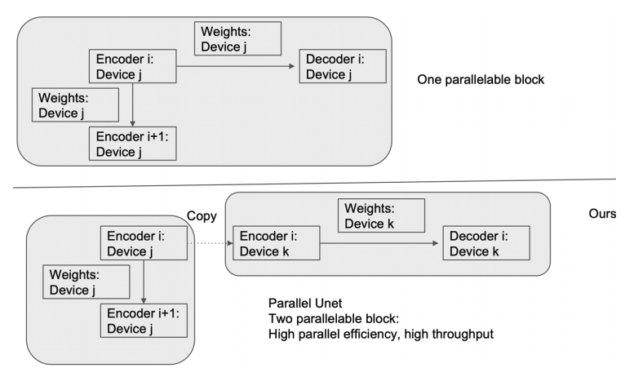
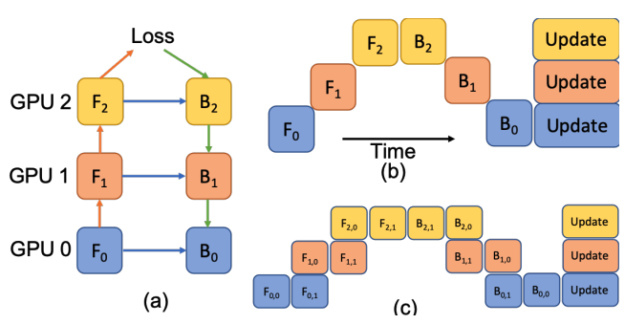
In Figure 5, a deep model is partitioned across three GPUs (a). Fk is the forward function of the k-th cell. Bk is the back-propagation function which relies on both Bk+1 from the upper layer and feature Fk. Conventional model parallelism has low device utilization because of dependency of the model (b). The pipeline parallelism splits the input minibatch to smaller micro-batches (c) and enables different devices to run micro-batches simultaneously. Synchronized gradient calculation can be applied last.
The MONAI research implementation shows straightforward implementations by using preprocessing modules such as the following:
AddChannelDictComposeRandCropByPosNegLabeldRand3DelasticdSpatialPadd
It also uses network modules, such as Convolution, and the layer factory to easily handle 2D or 3D inputs using the same module interface. The loss and metrics modules make the model training and evaluation simple. This implementation also includes a working example of training and validation pipelines.
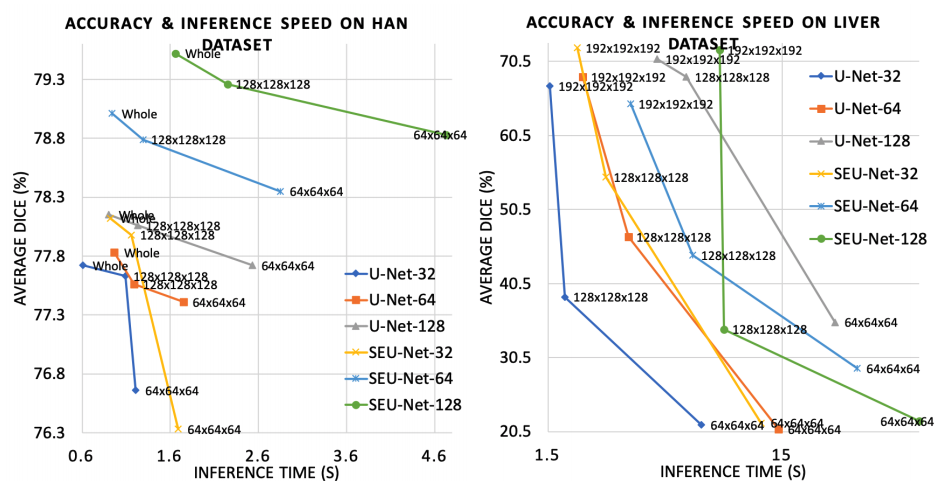
This research demonstrates the following:
- A large model and input increases segmentation accuracy.
- The large input reduces inference time significantly. LAMP can be a useful tool for medical image analysis tasks, such as large image registration, detection, and neural architecture search.
Summary
This post highlights how deep learning research for medical imaging could be built with MONAI. Both research examples use the representative features from MONAI v0.2.0, which allows for fast prototyping of research ideas.
For more information about MONAI v0.2.0, see the following resources: