Over the past couple of years, NVIDIA and NASA have been working closely on accelerating data science workflows using RAPIDS, and integrating these GPU-accelerated libraries with scientific use cases. This is the second post in a series that will discuss the results from an air pollution monitoring use case conducted during the COVID-19 pandemic, and share code snippets to port existing CPU workflows to RAPIDS on NVIDIA GPUs. This first post of this series, we covered Accelerated Simulation of Air Pollution.
Monitoring the Decline of Air Pollution Across the Globe During the COVID-19 Pandemic
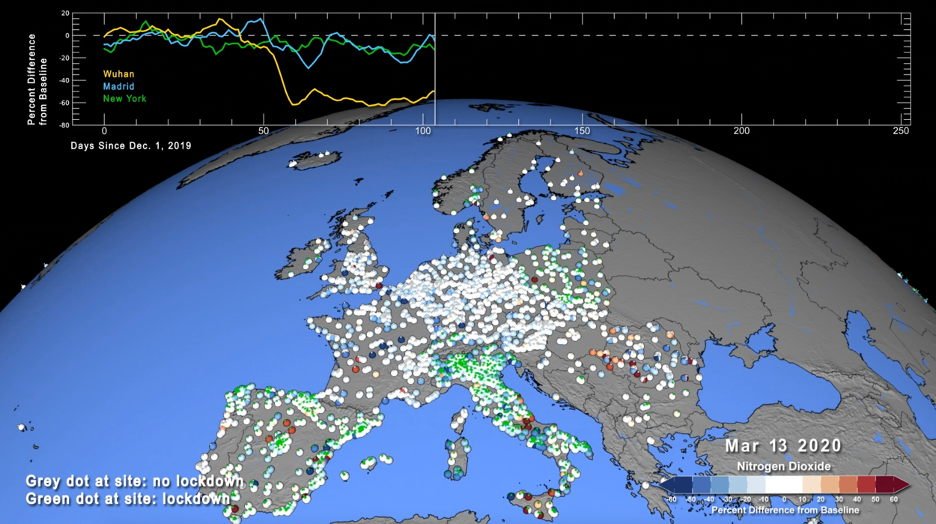
Another air quality application leveraging XGBoost and RAPIDS is the live monitoring of air quality through the combination of surface monitoring data and near real-time model data produced by the NASA GEOS-CF model. This approach is particularly useful to detect and quantify air pollution anomalies, i.e., patterns in air quality observations that cannot be explained by the model. The most prominent (and extreme) example of this is the decline of air pollution in the wake of the COVID-19 pandemic. As a result of the stay-at-home orders, traffic emissions of air pollutants such as nitrogen dioxide (NO2) decreased significantly, as apparent from both satellite observations and surface monitoring data. However, exactly quantifying the impact of COVID-19 restrictions on surface air quality solely based on these atmospheric observations is very difficult given that many other factors impact surface air pollution, including weather, chemistry, or wildfires.
The study conducted by Christoph and his colleagues fuses millions of observations – taken at 4,778 monitoring sites in 47 countries – with co-located model output produced by GEOS-CF. The sample code found at the covid_no2 repo, demonstrates the application for 10 selected cities (New York, Washington DC, San Francisco, Los Angeles, Beijing, Wuhan, London, Paris, Madrid, and Milan). The air quality observations for years 2018 through 2020 at these cities were obtained from the OpenAQ database (https://openaq.org/#/) and the European Environment Agency EEA (https://discomap.eea.europa.eu/map/fme/AirQualityExport.htm) and pre-processed into a single file for convenience:
Similarly, we preprocessed the GEOS-CF model output by subsampling the gridded native output (available in netCDF format at https://portal.nccs.nasa.gov/datashare/gmao/geos-cf/v1/das/) to the observation locations and saved the corresponding data as a table in text format that can be read similarly to the observation data:
The model data contains not only model predicted NO2 concentrations but also a number of ancillary model variables, such as information about the local weather and atmospheric composition (as taken from GEOS-CF) or calendar information.
The model data is then combined with the surface observation data to build an XGBoost bias-correction model that relates the model NO2 prediction to the observations. This is to account for the fact that the model prediction can be systematically different from the observations, e.g., because the model output represents the average over a 25×25 km2 domain while the surface observation is typically much more local in nature.
To train the XGBoost model, the model data is merged with the observations and the model bias is calculated from the merged data set to provide the label for the training (see sample code in repository mentioned above for full example):
Using the trained model, we can also calculate the SHAP values on GPU to analyze the factors that contribute most to the bias correction:
As shown in the figure below, the SHAP values for New York indicate that the most important predictors for the NO2 model bias (relative to the actual observation) is NO2 itself, followed by the hour of the day, wind speed (V10M) and the height of the planetary boundary layer (ZPBL). (note: to output the SHAP values in the example code the input argument shap needs to be set to 1, as well as gpu argument for accelerated code on the GPU).
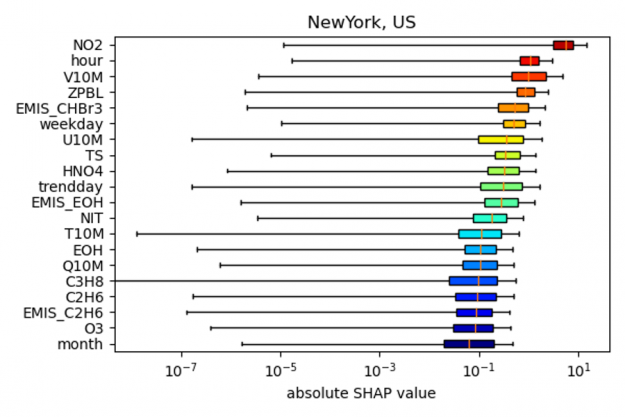
the bias correction model for New York City.
After training the XGBoost model on the 2018 and 2019 data (using 8-fold cross-validation), we extend the NO2 bias correction to the model data produced for year 2020, resulting in a time series of the expected NO2 concentrations at a given observation site if there had been no mobility restrictions due to the pandemic (the NO2 ‘baseline’). The difference between the actual observations and these bias-corrected model predictions offers an estimate of the impact of COVID-19 restrictions on NO2 concentrations. The figure below shows the difference between observations and model predictions at New York City from Jan 2019 to Jan 2021. The solid green line shows the best estimate, defined as the 21-day rolling average across all four observation sites available for New York City, and the dark and light shaded areas show two uncertainty estimates derived from the time-averaged and hourly model-observation samples, respectively.
Throughout year 2019, the bias-corrected model mean estimate is in close agreement with the observations. Coinciding with the outbreak of the pandemic, the observed NO2 over New York City declines by up to 40% and only gradually recovers to the expected value by year end.
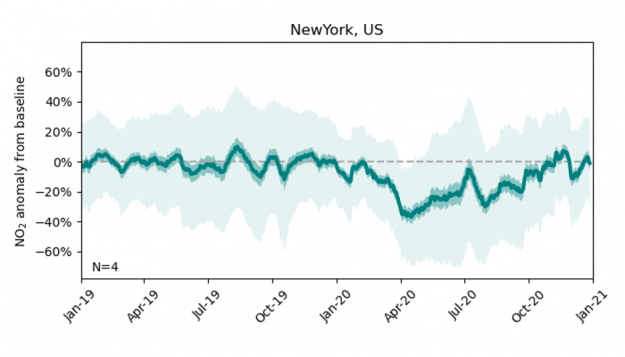
Figure 3: Dark and light-shaded areas indicate low and high uncertainty estimates.
Conducting this analysis at 4,778 locations across the world enables us to identify regional patterns in air quality anomalies, and our study shows that these patterns tend to be related to differences in timing and intensity of COVID-19 restrictions.
The here described approach is not only useful to analyze the impact of COVID-19 on air pollution but can be generally used to monitor air pollution across the world (both the observations and model data are available in near real-time). Given the growing number of available air quality observations, fast data processing becomes ever more critical for such an application. The sample code available at https://github.com/GEOS-CF/covid_no2 demonstrates that conducting the analysis on a V100 GPU using cuDF offers an overall speed-up of up to 5x for, each city compared to 20-core Intel Xeon E5-2689 CPU.
References:
Keller, C. A., Evans, M. J., Knowland, K. E., Hasenkopf, C. A., Modekurty, S., Lucchesi, R. A., Oda, T., Franca, B. B., Mandarino, F. C., Díaz Suárez, M. V., Ryan, R. G., Fakes, L. H., and Pawson, S.: Global impact of COVID-19 restrictions on the surface concentrations of nitrogen dioxide and ozone, Atmos. Chem. Phys., 21, 3555–3592, https://doi.org/10.5194/acp-21-3555-2021, 2021.
NASA Model Reveals How Much COVID-related Pollution Levels Deviated from the Norm